”I am inspired by the biological phenomena in which chemical forces are used in a repetitious fashion to produce all kinds of weird effects.” - Richard Feynman
Introduction
Today, there are a variety of devices which we can use to store digital data, like text, photos and videos. Wellknown examples include USB flash drives, hard disks, CDs, DVDs and Blu-Rays. We are also aware of a few obsolete storage devices like floppy disks and radio tapes which we do not use anymore. We are satisfied with the technology we have at the moment in terms of storing large volumes of digital data, and they have made fast-paced progress. Nevertheless, do we have to think of finding an alternative and a much efficient way to store data? Unfortunately, it turns out that these devices are approaching their density limits. In other words, the amount of data that these devices can store per unit volume are limited and fixed. As data and information are being generated in an exponential manner, it is expected that today’s storage devices might also be outdated soon. The information stored on them can only last for a few decades. This brings us to a situation where we have to hunt for a novel way to store large amounts of data for a very long period of time.
What’s so special about DNA?
DNA, or deoxyribonucleic acid, as we know is the genetic material present in most living organisms. They are double-helical structures consisting of four nucleotide bases - Adenine (A), Guanine (G), Cytosine (C) and Thymine (T) - along with sugar molecules and phosphate groups. The half-life of DNA is around 500 years in appropriate conditions, which means that it takes 500 years for just half of its bonds to break, making it highly stable. Interestingly, extensive research is taking place on using DNA to store digital data. Data is stored in electronic devices in the form of binary digits, 0 and 1. A simple idea is to map the sequence of binary digits to nucleotide sequences containing the four bases. One way to implement this is to map 2-bit sequences to the four nucleotides, as shown below.
| 00 | 01 | 10 | 11 |
| A | C | G | T |
In this way, we can convert large sequences of binary digits to sequences of nucleotide bases.
A brief history of DNA data storage
The concept of using DNA for data storage dates back to mid-1960s when the Soviet physicist Mikhail Neiman and the mathematician Norbert Wiener expressed their ideas of storing and retrieving digital data at the molecular level. In 1988, an artist named Joe Davis was able to encode a 35-bit image of an ancient Germanic rune representing life and the female Earth in a DNA sequence, which was then inserted into E. coli. In 1999, secret messages were hidden in DNA microdots on paper.
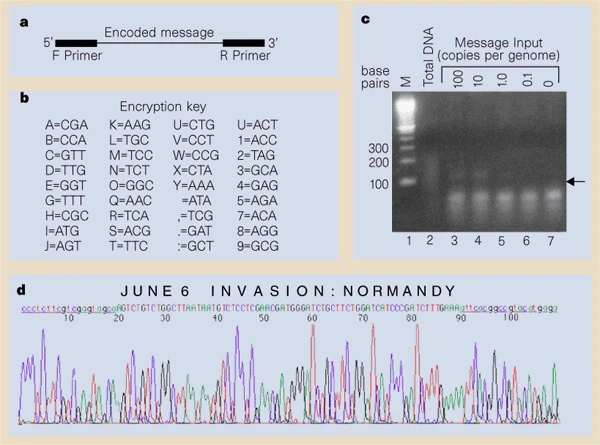
Figure 1: Genomic steganography: Secret messages encoded in DNA Source: https://www.nature.com/articles/21092
Following these remarkable attempts, several trials have been performed to improve the capacity and efficiency of DNA-based storage devices. More recently, in 2019, scientists were able to encode all of Wikipedia, totalling 16 GB, onto DNA strands.
A broad overview of the steps required
The four major steps involved in DNA data storage are: write, store, retrieve and read.
Writing and Storing
As mentioned previously, this step involves mapping strings of bits into DNA sequences by making use of computational algorithms. The binary sequences are usually broken down into smaller chunks and indices are added to each chunk to enable reassembly. The resulting DNA sequences are then synthesised. There are a variety of ways to synthesise DNA chemically. The most commonly used method is phosphoramidite-based oligonucleotide synthesis. The synthesised DNA sequences are stored in a library of DNA pools.
Retrieving and Reading
If we have to retrieve a particular data item from the library of DNA pools, we have to make sure that we choose that specific item among others rather than reading all the available data. This principle is called random access and is widely incorporated in computer memory. This can also be achieved in DNA storage by making use of PCR (Polymerase Chain Reaction) associated primers or magnetic bead extraction. Once the required data item is identified, we have to sequence it and convert it back to binary format. Again, there are a variety of ways to sequence DNA. Next-Generation Sequencing (NGS) is a massively parallel sequencing technique which is widely used on a commercial scale. Other methods include Sanger sequencing and Nanopore sequencing.
What about errors?
Both sequencing and synthesis are prone to errors like substitution, insertion and deletion of bases. In order to address these issues, error-correcting codes are implemented during the process of encoding. For example, redundant information is added to increase the probability that the original information can be retrieved even in the presence of errors and missing data. There are several ways to incorporate redundant information. Reed-Solomon codes are popular error-correcting codes that are also used to correct errors in present digital storage devices.
Storing data inside living cells!
Advancements in the field of synthetic biology have led to the construction of complicated synthetic genetic networks that can perform logical and state-dependent functions. Recording and storing of data can be done inside cells by using either a recombinase-based approach or CRISPR-based approaches. These systems can be used as biological recording devices which are more suited to collecting new data.
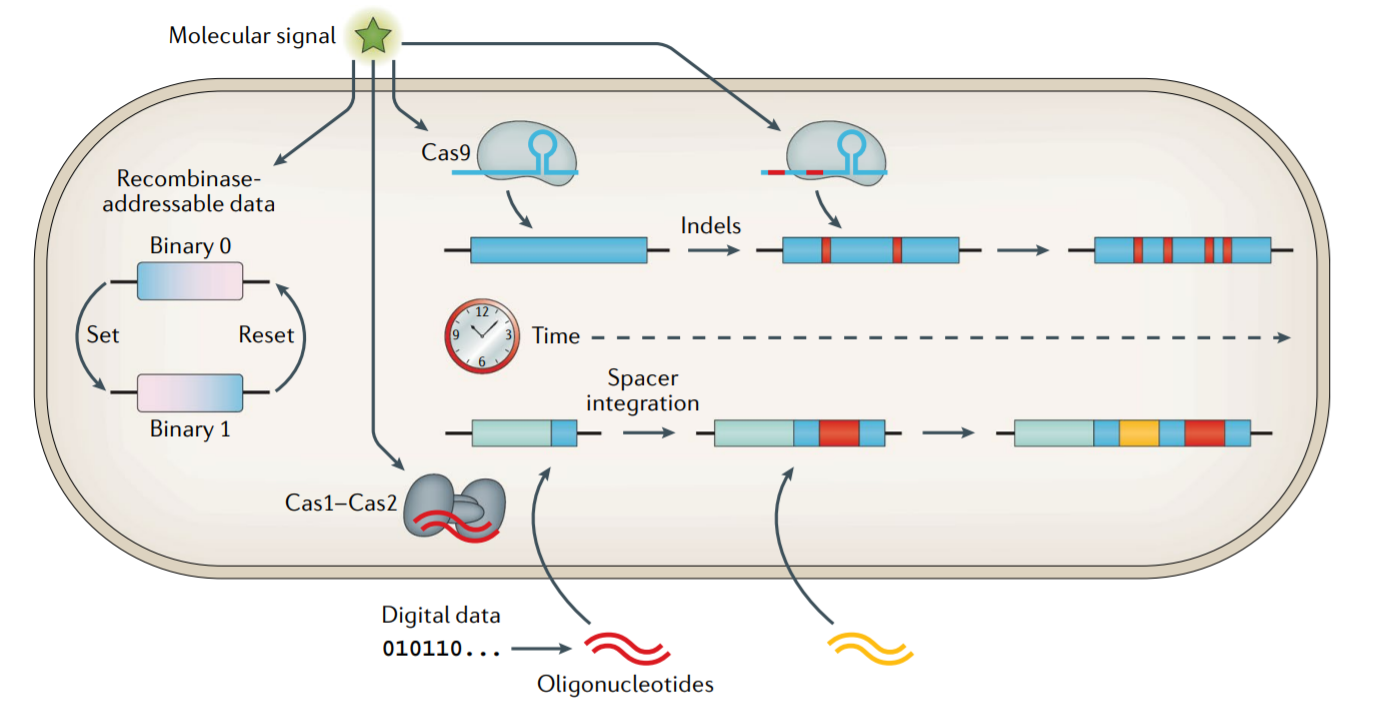
Figure 2: Storing and recording data in DNA - in vivo approach Source: https://www.nature.com/articles/s41576-019-0125-3
A Promising Alternative?
Despite the advantages of utilizing DNA to store data efficiently, there are several challenges and drawbacks that might need to be addressed. For example, it can take plenty of time to write and retrieve data, unlike modern data storage devices. Additionally, the degradation of DNA can be prevented only if it is stored under suitable conditions - low temperature and low humidity in the absence of ultraviolet rays. It is also important to develop better error-correcting mechanisms to improve the accuracy of the information obtained from the data. Hence, tremendous efforts are required to tackle these inherent problems.
Nevertheless, there is immense scope for DNA being the ideal storage device in the future due to its capacity to store enormous amounts of data in low volumes. It is fascinating to note that, theoretically, all the available data in the world can be stored in just a few hundred grams of DNA. Moreover, the ease of replication of DNA allows copying of data in constant time regardless of the size of the data. On the other hand, the copying time of any modern digital storage device is proportional to the size of the data.
In conclusion, as long as life exists on our planet, DNA can offer an eternal form of data storage that will never become obsolete!
Written by
Reference
Ceze, L., Nivala, J. & Strauss, K. Molecular digital data storage using DNA. Nat Rev Genet 20, 456–466 (2019). https://doi.org/10.1038/s41576-019-0125-3